David R. Stirling
We have now released CellProfiler Analyst version 3.0! Download it here.
As with CellProfiler 4, the primary goal of this release has been to migrate from Python 2 to Python 3 and modernise the application. We’ve also focused on improving performance and usability. Along with backend improvements that make the software faster and more reliable (see Performance), we’ve made some significant changes to how you can easily and efficiently interact with the software (see Interface Refinements) and your data (see New Functionality).
New Functionality
We’ve added some exciting new features to CellProfiler Analyst:
- Added the “Dimensionality Reduction” tool. This plot interface allows you to condense high-dimensional data sets (those featuring many measurements) into a smaller set of components reflecting overall variance. This can be useful for identifying outliers and other groups of interest within your data. The available reduction methods include PCA, SVD, Factor Analysis, t-SNE, and several others. See the manual for more details.
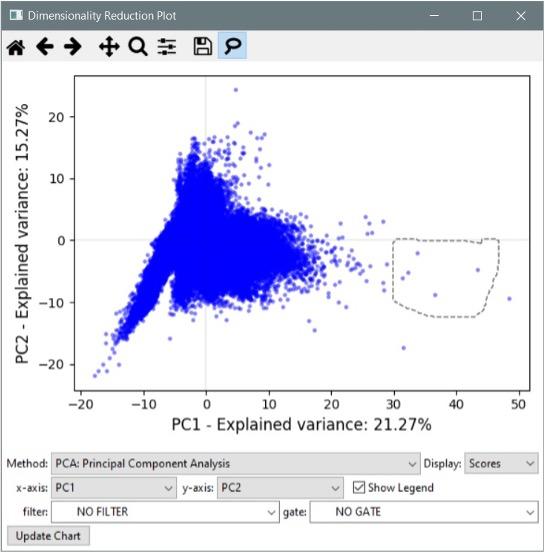
- The dimensionality reduction plots have a “lasso” tool for selecting objects. Draw around objects to select them, then right click to see options for displaying them. If a classifier window is open, you can send the selected objects directly to the classifier.
- New classifier type: Neural Network. You can now classify objects using customisable neural networks. These can be particularly useful for performing complex non-linear classification tasks.
- Classifier models now support scaling to normalise data before classification; this can be toggled on/off in the “advanced” menu. Scaling is enabled by default on model types which most benefit from it (SVC, KNeighbours and Neural Network).
- Filters can now operate at the per-object level, rather than being limited to operating on whole images. This enables you to visualise more specific populations of interest within the plotting tools.
- Gates are now directly available in the classifier. No need to add them to the .properties and restart CellProfiler Analyst.
- You can now fetch objects from an image in sequential order within the classifier, instead of sampling randomly. This can be useful when working to classify objects in a specific order, as determined by your filters.
Interface refinements
We’ve made refinements to CellProfiler Analyst’s interface to address issues commonly raised by the community. Some major changes are as follows:
- Image loading and tile manipulation is now much smoother. Dragging multiple tiles should no longer lock up the program.
- You can now switch properties files without restarting CellProfiler Analyst by using the File menu on the main window.
- Properties file errors will now be caught and displayed to you, rather than crashing CellProfiler Analyst.
- Within the classifier, you can now drag to select multiple tiles at once.

- Within the classifier, you can now use the arrow keys and number keys to select tiles and move them into class bins. E.g. pressing “1” will move any selected tiles from the “Unclassified” bin into the first class that you defined, thus enabling rapid classification using only the keyboard.
- You can now optionally prevent duplicate objects in the Classifier by using the “Advanced” menu and remove existing duplicates with the right-click menu within each bin. Randomly sampling objects can return the same object multiple times. Sometimes this is useful for reinforcing training, but users have often requested the ability to suppress these duplicates.
- A “fit to window” button is now available in the image viewer.
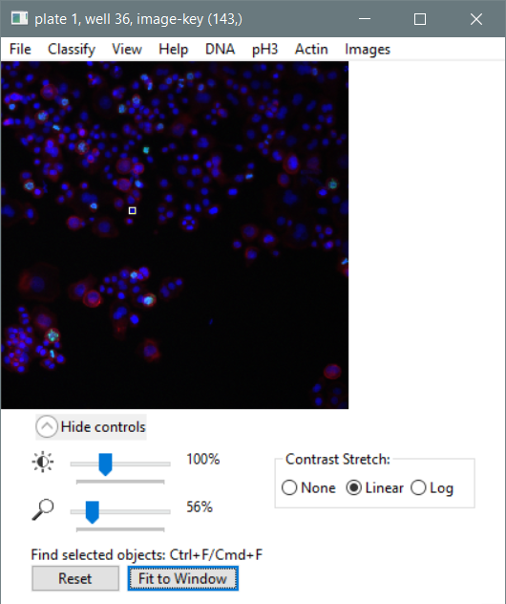
- The image size/contrast control panel now only updates the display when the slider stops moving. This should prevent CellProfiler Analyst from freezing when the user tries to adjust display settings with multiple tiles already loaded.
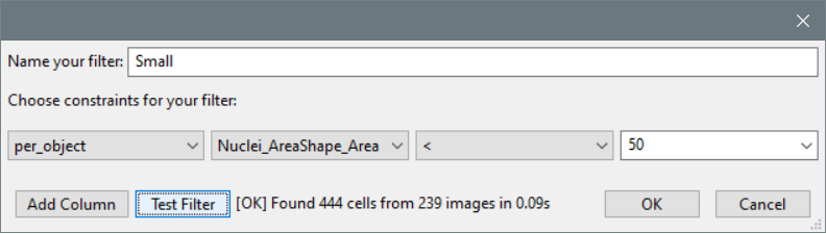
- The “Create Filter” dialog now includes a “Test” button so that filters can be validated before saving them.
Performance
Another area of focus has been the performance of different aspects of CellProfiler Analyst. Some improvements include:
- The “Score Image” and “Score All” functions within the classifier have been revised to run more efficiently. Scoring should now be much faster: In one of our testing datasets, scoring 50,000 objects with a RandomForest classifier completed in ~20 seconds,compared to over 10 minutes in CellProfiler Analyst 2.
- Handling of custom SQL filters has been optimised.
- We’ve made more general improvements to database interactions to minimise the number of SQL calls which are made. This particularly impacts random sampling of objects with the classifier and graphing tools. In most cases operations will be much faster, but if users encounter issues they can revert to the old system by using the “use_legacy_fetcher = True” properties file option.
- Loading saved training sets is now substantially faster.
- The methods for drawing image tiles have been improved, particularly when running CellProfiler Analyst in whole-image classification mode.
Compatibility Notes
This is a major release, and so this version introduces changes to CellProfiler Analyst that will change behaviour compared to previous versions. Some points to note:
- Classifier .model files can be exported, although note that they will only be compatible with the next release of the companion software CellProfiler 4 (4.1.3 should be the last incompatible version). They will not work at all with the CellProfiler 3 series due to the change in programming language from Python 2 to Python 3. Outside of CellProfiler, models can be loaded in a Python environment using joblib and scikit-learn. If enabled, the scaler normalization function is attached to the saved model as model.scaler, which can be used to transform new input data.
- CellProfiler Analyst’s Java dependencies are now packaged with built versions of the application; installing a separate JDK is no longer required.
- The ImageIO package is now used as the default image reader in CellProfiler Analyst. This reader is generally much faster at loading files, but does not support as many file formats. Bioformats will be used for formats not compatible with ImageIO. You can revert to using only Bioformats by adding the “force_bioformats = True” flag to your .properties file.
Contributors
The following people contributed to the CellProfiler Analyst 3.0 release — we’re very grateful for all their help!
David Stirling, Pearl Ryder, Beth Cimini and Jane Hung